TimeXNet is an application that predicts activated pathways during a cellular response using time-course gene expression profiles in the context of a large molecular interaction network.
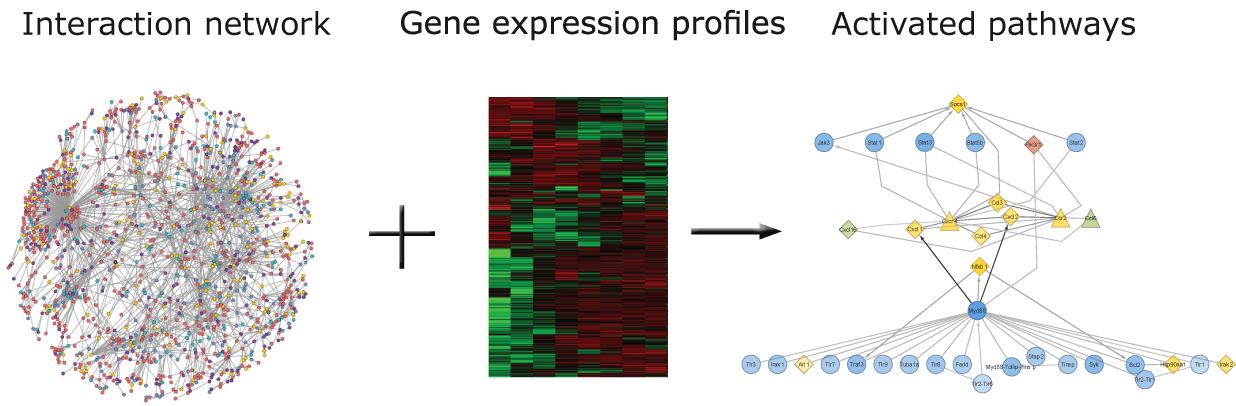
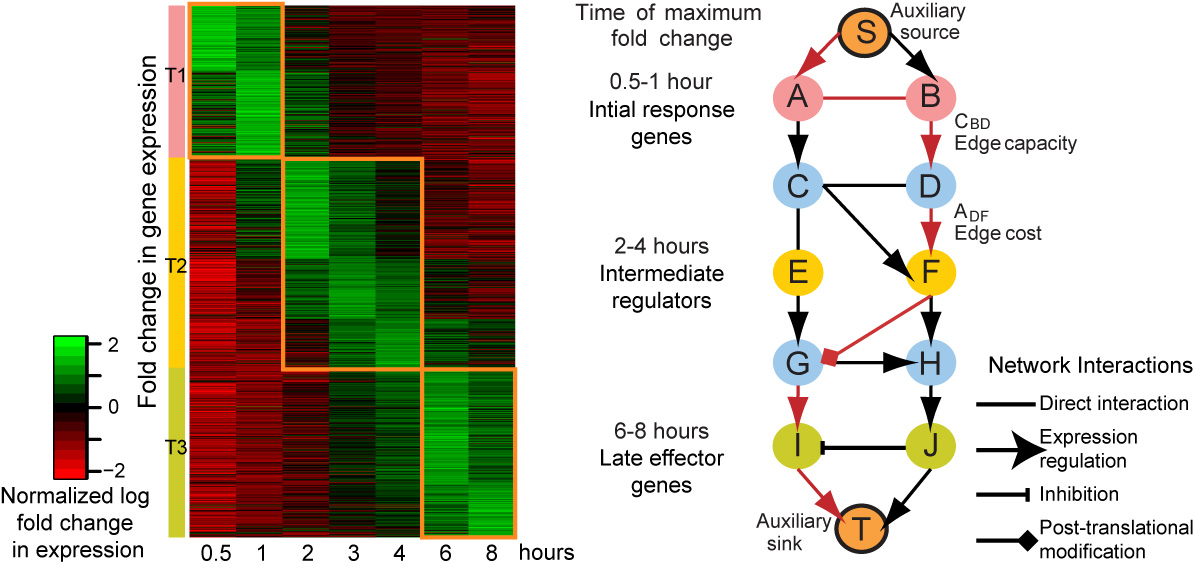
TimeXNet implements an algorithm based on a minimum cost flow optimization approach to predict activated pathways in a large molecular network between three sets of genes – initial, intermediate and late response genes based on their time of expression.
TimeXNet is implemented in Java and can be run on the command-line or through a user-interface. It requires two main sources of information:
-
Three groups of genes partitioned by the time of their highest fold change in expression, and scored based on their expression levels
-
A large molecular network containing protein-protein, protein-DNA interactions and post-translational modification with edges weighed according to their reliability
TimeXNet was used to identify the activated pathways during the innate immune response in mouse dendritic cells stimulated with LPS. For more details, please refer to:
Linking Transcriptional Changes over Time in Stimulated Dendritic Cells to Identify Gene Networks Activated during the Innate Immune Response. Patil A, Kumagai Y, Liang KC, Suzuki Y, Nakai K. PLoS Comput Biol. 2013 Nov;9(11):e1003323. PDF